CountBio
Mathematical tools for natural sciences
Basic Statistics with R
Welsch ANOVA
In one way ANOVA, the independent samples are assumed to be drawn from Gaussian distributions whose variances are equal.
When this assumption on the equality of the variances across samples is not met, Welsch ANOVA (also called "Welsch F-test")
is a better option for testing the equlaity of more than two populations means.
This method still assumes that the data sets whose population means are compared are randomly drwn from Guassians with unequal variances.
Thus the Welsch ANOVA is an alternative to one factor ANOVA when the variances are unequal or sample sizes are small.
In Welch F-test, a weight $w_i$ which is inversely proportional to the variance of sample $i$ is used to reduce the effect of
heterogenity across the samples.
If \(s_i\) is the observed variance of sample \(i\), then the weight \(w_i\) for the sample of size $n_i$ is defined as,
\(~~w_i = \dfrac{n_i}{s_i^2} \)
The rest of the procedure is very similar to one way ANOVA except for the fact that the weighted means and variances are used instead of
normal means and variances.
In the following description, we adopt the same methods and symbols we have used in our derivation of one way anova.
Thus, in Welsch F-test, the grand mean $\overline{X}_{..}$ of the whole data is computed based on the weighted mean for each data set.
If there are m data sets, then the grand mean of the Welsch ANOVA is written as,
$~~~~~~~~~~~~~~\overline{X}_{..}~=~\displaystyle { \dfrac{\sum\limits_{i=1}^m w_i \overline{X}_{i.}}{\sum\limits_{i=1}^{m} w_i}} ~~~~~~~~~$
where $\overline{X}_{i.}$ is the sample mean of the ${\large{i}}^{th}$ group.
In the next step, using the weights, we compute the sum of squares among the treatements (called between the sum of square) as,
$~~~~~~~~~~~~~SSA_{w}~=~\displaystyle \sum\limits_{i=1}^m w_{i}(\overline{X}_{i.} - \overline{X}_{..})^2 $
We denote $ w = \displaystyle \sum\limits_{i=1}^{m} w_{i}$ and introdyce a variable $\lambda$ as,
$~~~~~~~~~~~~~~~{\lambda} = \displaystyle \dfrac{3 \sum\limits_{i=1}^{m} \dfrac{1}{(n_i - 1)} \left(1 - \dfrac{w_i}{w}\right)^2 } {m^2 - 1} $
Using this variable, the Welsch F statistic for the statistical test is defined as,
$~~~~~~~~~~~~~~F_w~=~\displaystyle \dfrac{ {\left(\dfrac{SSA_w}{m-1}\right)} }{1 + \dfrac{2\lambda(m-2)}{3} }~~~{\Large \sim}~~~ F(m-1, \dfrac{1}{\lambda})~~$ is a F variable with $(m-1, \dfrac{1}{\lambda})$ degrees of freedom under the null hypothesis of equality of means.
The null and alternate hypothesis for the Welsch ANOVA are written as,
$~~~~~~~~~~~~~H_0~:~~\mu_1 = \mu_2 = \mu_3 = ...... = \mu_m $
$~~~~~~~~~~~~~H_A~:~~Not~all~the~\mu_i~are~equal$.
Therefore, if $\alpha$ is the significance level for rejecting the test,
We accept the null hypothesis $H_0~$ if $~ F_w \leq F_{1-\alpha}(m-1, \dfrac{1}{\lambda})$
We reject the null hypothesis $H_0$ and accept the alternative if $~ F_w \gt F_{1-\alpha}(m-1, \dfrac{1}{\lambda})$
$~~~~~~~~~~~~~~~~~~~~~~~~~$ Important Note :
When the assumptions on the equality of mean are not met, we can use Welsch ANOVA instead of one way ANOVA. However, this method
has two limitations:
$~~~~~~$ 1. Welsch ANOVA has fewer degrees of freedom when compared to one way ANOVA since $\dfrac{1}{\lambda} \leq (n + m -1)$, where m is the number of data sets and n is the total number of data points and (n+m-1) is the degrees of freedom used in the one way ANOVA.
Cosequently, Welsch ANOVA is less powerful than the one way ANOVA test.
2. Since the weight factor in the Welsch ANOVA is $w_i = \dfrac{n_i}{s_i^2}$, when the number of obdervations $n_i$ are small, the corresponding weight factor gets smaller making the Welsch ANOVA less reliable than the one way ANOVA for the small sample sizes.
Example-1 :
The cholesterol level of four groups of adults with distince food habits among the groups were compared in an experiment. The results are present below:
$Group-1~:~~220~~214~~203~~184~~186~~200~~165$
$Group-2~:~~262~~193~~225~~200~~164~~266~~179$
$Group-3~:~~272~~192~~190~~208~~231~~235~~141$
$Group-4~:~~190~~255~~247~~278~~230~~269~~289$
Assuming that these four data sets follow Normal distributions $N(\mu_i, \sigma)$, test the null hypothesis that $\mu_1=\mu_2=\mu_3=\mu_4$ to a significance level of 0.05.
We assume that the four populations to be Gaussian with unequal standard deviations and perform Welsch ANOVA to test the equality of their populations means.
For the convenience of symbols, let us denote the four data sets as $X_1, X_2, X_3$ and $X_4$. We write the ANOVA table to perfor the computation as follows:
------------------------------------------------------------------------------------------------------------------------------
$Variable~~~~~~~~~~~~~~~~~~~~~Observations~~~~~~~~~~~~~~~~~~~~~~n_i~~~~~~~\displaystyle\sum_{j=1}^{7}X_{ij}~~~~~~~S_i^2~~~~~~~~~~~~w_i = \dfrac{n_i}{s_i^2}$
------------------------------------------------------------------------------------------------------------------------------
$~~~X_1~~~~~~~~~~~~220~~214~~203~~184~~186~~200~~165~~~~~~~~~7~~~~~~~~1372~~~~~~~~~~~361.66~~~~~~~~~0.019350$
$~~~X_2~~~~~~~~~~~~262~~193~~225~~200~~164~~266~~179~~~~~~~~~~7~~~~~~~~1489~~~~~~~~~~1579.90~~~~~~~0.004430$
$~~~X_3~~~~~~~~~~~~272~~192~~190~~208~~231~~235~~141~~~~~~~~~~7~~~~~~~~1469~~~~~~~~~~1733.14~~~~~~~0.004039$
$~~~X_4~~~~~~~~~~~~190~~255~~247~~278~~230~~269~~289~~~~~~~~~~7~~~~~~~~1758~~~~~~~~~~1115.14~~~~~~~0.006277$
--------------------------------------------------------------------------------------------------------------------------------
$~~~~~~~~~~~~~~~~~~~~Totals~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~28~~~~~~~~~6088~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~0.03410$
--------------------------------------------------------------------------------------------------------------------------------
We now compute the mean $\overline{X}_{i.}$ each group i and the whole mean $\overline{X}_{..}$ as follows:
$~~~\overline{X}_{1.} = \dfrac{1372}{7} = 196.0$, $~~~\overline{X}_{2.} = \dfrac{1489}{7} = 212.7$,
$~~~\overline{X}_{3.} = \dfrac{1469}{7} = 209.85$, $~~~\overline{X}_{4.} = \dfrac{1759}{7} = 251.14$
$\overline{X}_{..}~=~\displaystyle { \dfrac{\sum\limits_{i=1}^m w_i \overline{X}_{i.}}{\sum\limits_{i=1}^{m} w_i}}$
$~~~=~\dfrac{(0.019350 \times 196.0) + ( 0.004430 \times 212.7) + (0.004039 \times 209.8) + (0.006277 \times 251.3)}{0.034096}$
$~~~=~~209.96$
We next compute the Sum of Squares among the treatment:
$SSA_{w}~=~\displaystyle \sum\limits_{i=1}^m w_{i}(\overline{X}_{i.} - \overline{X}_{..})^2 $
$~~~~~~~~~~=~0.019350 \times (196.0 - 209.98)^2 ~+~ 0.004430 \times (212.7 - 209.98)^2$
$~~~~~~~~~~~~~~~~~~~~~~~+~ 0.004039 \times (209.8 - 209.98)^2~+~0.006277 \times (251.3 - 209.98)^2 $
$~~~~~~~~~~=14.4519$
The sum of weights$~~ w = \displaystyle \sum\limits_{i=1}^{m} w_{i}~=~ 0.034096,~~~$ number of groups $~~m = 4$
.
${\lambda} = \displaystyle \dfrac{3 \sum\limits_{i=1}^{m} \dfrac{1}{(n_i - 1)} \left(1 - \dfrac{\omega_i}{\omega}\right)^2 } {m^2 - 1} $
$~~~=~ \dfrac{3}{15} \times \left[ \dfrac{1}{6} \left(1 - \dfrac{0.019350}{0.034096}\right)^2 + \dfrac{1}{6} \left(1 - \dfrac{0.004430}{0.034096}\right)^2
+ \dfrac{1}{6} \left(1 - \dfrac{0.004039}{0.034096}\right)^2 \\ ~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~+ \dfrac{1}{6} \left(1 - \dfrac{0.006277}{0.034096}\right)^2 \right ] $
$~~~~~=~0.07956$
$~~~~~~~~~~~~~~F_w~=~\displaystyle \dfrac{ {\left(\dfrac{SSA_w}{m-1}\right)} }{1 + \dfrac{2\lambda(m-2)}{3} }~=~\displaystyle \dfrac{ {\left(\dfrac{14.4519}{4-1}\right)} }{1 + \dfrac{2 \times 0.07956 \times(4-2)}{3} } ~= 4.3552$
This $F_w$ is a F variable with $(m-1, \dfrac{1}{\lambda})$ degrees of freedom.
Here,$~~~~~~m-1 = 3,~~~\dfrac{1}{\lambda} = \dfrac{1}{0.07956}~=~12.569$
$F_{1 - 0.05}(3,12.569)~=~qf(0.95,3,12.56)~=~3.4437$
The critical region for rejection of null hypothesis is $~~F \ge 3.4437$.
Since the computed value $F=4.3552$ is outside the critical region, we reject the null and accept the alternate hupothesis to conclude that the four population means are not equal.
The p-value for the test is given by area under
$F(>4.3552, 3, 12.56) = 1 - pf(4.3552, 3, 12.569) = 0.025766$,
which rejects the null at a significance level of 0.05.
R scripts
In R, two functions namely aov() andoneway.test() perform one way anova. They have a very specific inut data structure in the form of a 2 column data frame with category and values of data. First step is to create the input file from data. Te second step is to run call the functions in R script. For example, consider the cholesterol level data among four groups Group-1, Group-2, Group-3 and Group-4 presented in Example-1. We show it below: Group−1 : 220 214 203 184 186 200 165 Group−2 : 262 193 225 200 164 266 179 Group−3 : 272 192 190 208 231 235 141 Group−4 : 190 255 247 278 230 269 289 We shorten the names to G1, G2, g3 and G4. These 4 setscan be written as a two column data in which first column is the value against which the group label is given. See below: (two colums can be tb separated(*.txt file) or comman separated (*.csv file) value group 220 g1 214 g1 203 g1 184 g1 186 g1 200 g1 165 g1 262 g2 193 g2 225 g2 200 g2 164 g2 266 g2 179 g2 272 g3 192 g3 190 g3 208 g3 231 g3 235 g3 141 g3 190 g4 255 g4 247 g4 278 g4 230 g4 269 g4 289 g4 The above data format is stored as a text file with a txt extension. We give some name like,"cholesterol_anova_data.txt" . It can also be stored as a comma separated csv format. This has the advantage that we can handle data sets of different lengths. If you store it as data frame, then many NA's may be required to make them equaal length. Above format is better. Once this file"anova_data.txt" is ready, we can write an R script for performing one factor ANOVA as follows:
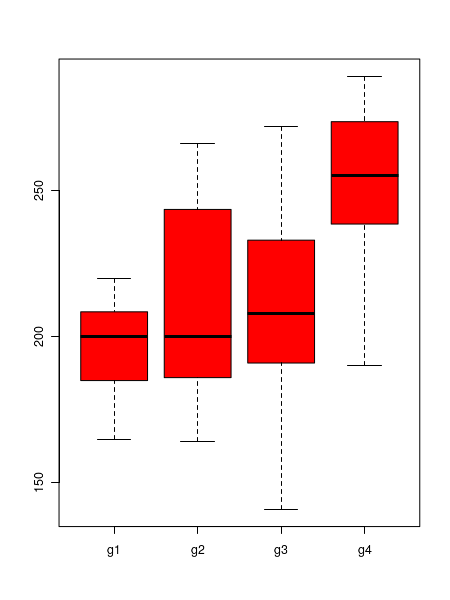
##analysis of oneway anova in R ## read data into a frame called "dat" dat = read.table("cholesterol_anova_data.txt", header=TRUE) ## compute and print the mean values of groups print( mean(dat$value[dat$group=="g1"]) ) print( mean(dat$value[dat$group=="g2"]) ) print( mean(dat$value[dat$group=="g3"]) ) print( mean(dat$value[dat$group=="g4"]) ) ## box plot to compare the data sets boxplot(dat$value ~ dat$group, col="red") # one way anova test. Default assumption is variances not equal. oneway.test(dat$value~dat$group, var.equal=FALSE)(i) In the above script, the callboxplot(dat$value ~ dat$group, col="red") creates the following plot: We can clearly see that there is a substantial difference of means within the four data sets. (ii) The function calloneway.test(dat$value~dat$group, var.equal=FALSE) with the assumption that the variances are unequal (default assumption, Welsch anova) to print the following results:For a F statisti value of 4.355 with 3 degrees of freedom, the p-value for the test is 0.02577. If we set an alpha = 0.05, this rejects the null hypothesis that the mean cholesterol levels for the 4 groups are equalOne-way analysis of means (not assuming equal variances) data: dat$value and dat$group F = 4.3553, num df = 3.000, denom df = 12.568, p-value = 0.02577